Question 1
How can fragments of DNA be separated?
A. Using polymerase chain reaction (PCR)
B. Using gel electrophoresis
C. Using gene transfer
D. Using gene cloning
Easy
Mark as Complete
Mark Scheme
Question 2
Which technique causes fragments of DNA to move in an electric field?
A. Polymerase chain reaction (PCR)
B. Genetic modification
C. Therapeutic cloning
D. Gel electrophoresis
Easy
Mark as Complete
Mark Scheme
Question 3
What is amplified using the polymerase chain reaction (PCR)?
A. Large amounts of RNA
B. Small amounts of DNA
C. Small amounts of protein
D. Large amounts of polymers.
Easy
Mark as Complete
Mark Scheme
Question 4
What happens to DNA fragments in electrophoresis?
A. They move in a magnetic field and are separated according to their size.
B. They move in an electric field and are separated according to their size.
C. They move in a magnetic field and are separated according to their bases.
B. They move in an electric field and are separated according to their bases.
Easy
Mark as Complete
Mark Scheme
Question 5
How can forensic scientists obtain sufficient data from one hair follicle to make a reliable identification by DNA profiling?
A. By performing PCR with DNA from the sample
B. By digesting the sample with more than one restriction enzyme
C. By performing electrophoresis with many other known samples
D. By choosing a hair follicle with a particularly long hair
Easy
Mark as Complete
Mark Scheme
Question 6
The diagram shows a gel electrophoresis apparatus.
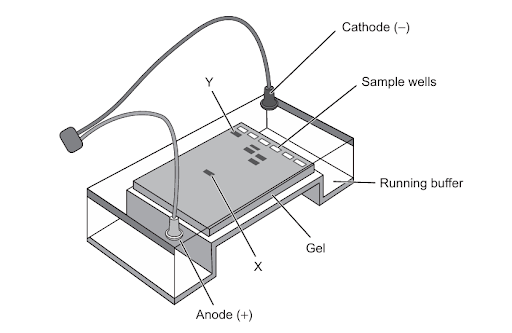
What can be deduced from the diagram?
A. X is the smallest fragment.
B. Y is positively charged.
C. X is moving towards the cathode.
D. Y travels at the greatest rate.
Medium
Mark as Complete
Mark Scheme
Question 7
What is the function of DNA polymerase I in DNA replication?
A. It forms primers by adding short lengths of RNA to the template strand.
B. It removes RNA primers and replaces them with DNA.
C. It builds the leading strand by adding DNA nucleotides continuously.
D. It forms Okazaki fragments by adding DNA nucleotides on the lagging strand.
Easy
Mark as Complete
Mark Scheme
Question 8
Which enzyme is associated with proofreading during DNA replication?
A. DNA primase
B. DNA helicase
C. DNA polymerase III
D. DNA ligase
Medium
Mark as Complete
Mark Scheme
Question 9
(a) Explain why DNA must be replicated before mitosis and the role of helicase in DNA replication. [4]
(b) Explain how the base sequence of DNA is conserved during replication. [5]
Hard
Mark as Complete
Mark Scheme
Question 10
(a) Draw a labelled diagram of the structure of DNA, showing the arrangement of subunits. [3]
(b) Explain DNA replication. [5]
Medium
Mark as Complete
Mark Scheme
Question 1
How can fragments of DNA be separated?
A. Using polymerase chain reaction (PCR)
B. Using gel electrophoresis
C. Using gene transfer
D. Using gene cloning
Answer: B
A. Incorrect: PCR technique for amplifying selected regions of DNA by multiple cycles of DNA synthesis - not to separate fragments. During PCR, the DNA is replicated repeatedly through cycles of heating and cooling using an enzyme called heat-tolerant DNA polymerase. Although PCR produces DNA fragments, it doesn’t separate them; that step comes after PCR.
B. Correct: Gel electrophoresis is a laboratory technique used to separate DNA fragments based on their size and charge. DNA fragments are placed in a polymer gel and an electric current is applied. Since DNA is negatively charged - because of the phosphate groups, fragments move toward the positive pole (anode). Smaller fragments move faster and travel farther, while larger fragments move slower.
C. Incorrect: Gene transfer means moving a genetic material from one organism to another. It is used in genetic engineering to introduce new genes into cells. It doesn’t separate DNA fragments.
D. Incorrect: Gene cloning involves making identical copies of a particular gene or DNA sequence, usually inside a host cell. It’s used to produce large amounts of a specific DNA or protein. While cloning also uses DNA manipulation, it doesn’t separate fragments - it replicates them.
Question 2
Which technique causes fragments of DNA to move in an electric field?
A. Polymerase chain reaction (PCR)
B. Genetic modification
C. Therapeutic cloning
D. Gel electrophoresis
Answer: D
A. Incorrect: PCR technique for amplifying selected regions of DNA by multiple cycles of DNA synthesis. It involves heating and cooling cycles, not an electric field. No movement of DNA fragments occurs - only replication using enzymes.
B. Incorrect: Genetic modification means altering an organism’s DNA, often by inserting new genes using vectors like plasmids. This process does not involve an electric field or movement of DNA fragments through a gel.
C. Incorrect: Therapeutic cloning involves creating an embryo from a somatic cell and using its stem cells for medical treatment. It does not deal with separating DNA or applying an electric current to DNA fragments.
D. Correct: Gel electrophoresis is the technique that uses an electric field to separate DNA fragments based on their size and charge. DNA molecules are negatively charged due to phosphate groups in their backbone. When placed in a polymer gel and exposed to an electric current, DNA fragments move toward the positive pole (anode).
Question 3
What is amplified using the polymerase chain reaction (PCR)?
A. Large amounts of RNA
B. Small amounts of DNA
C. Small amounts of protein
D. Large amounts of polymers.
Answer: B
A. Incorrect: The polymerase chain reaction (PCR) works on DNA, not RNA. In PCR, Taq polymerase cannot synthesize RNA because it doesn’t recognize RNA nucleotides and isn’t built to make RNA strands. RNA must first be converted into complementary DNA (cDNA) by reverse transcriptase before it can be amplified, which called Reverse Transcription PCR (RT-PCR).
B. Correct: In the polymerase chain reaction (PCR), double-stranded DNA is amplified, meaning many copies are made. Often, it is only possible to produce or recover a very small amount of DNA (such as in genetic engineering or at a crime scene). In PCR, DNA is replicated in an entirely automated process, in vitro, to produce a large amount of the sequence.
C. Incorrect: PCR does not involve proteins; it only works on nucleic acids (DNA). Proteins are studied using different methods such as western blotting or mass spectrometry
D. Incorrect: The term “polymer” here is misleading. DNA is a type of polymer, but PCR does not amplify all polymers - only specific DNA sequences. “Large amounts of polymers” is too general and scientifically inaccurate.
Question 4
What happens to DNA fragments in electrophoresis?
A. They move in a magnetic field and are separated according to their size.
B. They move in an electric field and are separated according to their size.
C. They move in a magnetic field and are separated according to their bases.
B. They move in an electric field and are separated according to their bases.
Answer: B
A. Incorrect: Electrophoresis uses an electric field, not a magnetic field. DNA fragments carry a negative charge due to their phosphate backbone. They move only when an electric current is applied, magnets don’t affect DNA. So, while separation does depend on size, the field type (magnetic) is wrong.
B. Correct: DNA is negatively charged, so in an electric field, it moves toward the positive pole (anode). The gel acts like a molecular sieve - small fragments move more easily and travel farther, while large fragments move more slowly. Therefore, separation occurs by fragment size.
C. Incorrect: Magnetic field isn’t involved in electrophoresis. DNA fragments are not separated by their bases (A, T, G, C) - every DNA molecule has the same overall charge-to-mass ratio. So neither part of this statement is correct.
D. Incorrect: The first part “electric field” is correct, DNA moves under an electric field. But the second part “separated according to their bases” is wrong. DNA fragments with different base sequences but the same length will move the same distance in the gel.
Question 5
How can forensic scientists obtain sufficient data from one hair follicle to make a reliable identification by DNA profiling?
A. By performing PCR with DNA from the sample
B. By digesting the sample with more than one restriction enzyme
C. By performing electrophoresis with many other known samples
D. By choosing a hair follicle with a particularly long hair
Answer: A
A. Correct: PCR (Polymerase Chain Reaction) is a technique that makes millions of copies of a small DNA sample. A single hair follicle contains very little DNA, not enough for profiling. By using PCR, scientists can amplify (multiply) the DNA many times, producing sufficient DNA for analysis. This allows reliable DNA profiling - even from a tiny or degraded sample like a single hair root.
B. Incorrect: Restriction enzymes are used to cut DNA into fragments at specific sequences, but they don’t increase the amount of DNA. Using more enzymes just creates more pieces, not more DNA. So, this does not solve the problem of limited DNA quantity.
C. Incorrect: Gel electrophoresis is a later step which is used to separate DNA fragments by size. Running electrophoresis with other samples may help in comparison, but it does not increase the DNA available from the hair follicle. Without enough DNA first (through PCR), electrophoresis cannot even be performed accurately.
D. Incorrect: The length of the hair shaft doesn’t affect the amount of nuclear DNA, DNA is only found in the follicle (root), not in the hair strand. A longer hair doesn’t mean more DNA for profiling. Therefore, the length is irrelevant to obtaining sufficient DNA.
Question 6
The diagram shows a gel electrophoresis apparatus.

What can be deduced from the diagram?
A. X is the smallest fragment.
B. Y is positively charged.
C. X is moving towards the cathode.
D. Y travels at the greatest rate.
Answer: A
A. Correct: In gel electrophoresis, DNA fragments are negatively charged and move from the cathode (–) to the anode (+) when an electric current is applied. Smaller DNA fragments move faster and farther through the gel because they can pass more easily through the pores of the gel matrix. In the diagram, X is farther from the wells, meaning it traveled the farthest. Therefore, X must be the smallest fragment, because small fragments move the greatest distance.
B. Incorrect: All DNA fragments are negatively charged, due to the phosphate groups in the backbone. They move toward the positive pole (anode), not away from it. So Y cannot be positively charged - if it were, it would move in the opposite direction (toward the cathode). DNA is always negatively charged, so this is wrong.
C. Incorrect: The cathode (–) is where DNA starts, near the sample wells. DNA moves away from the cathode and toward the anode (+) because it is negatively charged. X is located near the anode, showing it has already moved away from the cathode. Direction is wrong - DNA moves toward the anode, not the cathode.
D. Incorrect: Y is closer to the wells, meaning it has traveled a shorter distance. Large DNA fragments move more slowly because they have more difficulty moving through the gel matrix. Since X is farther along the gel, X travels faster, not Y. Y travels the slowest, not the fastest.
Question 7
What is the function of DNA polymerase I in DNA replication?
A. It forms primers by adding short lengths of RNA to the template strand.
B. It removes RNA primers and replaces them with DNA.
C. It builds the leading strand by adding DNA nucleotides continuously.
D. It forms Okazaki fragments by adding DNA nucleotides on the lagging strand.
Answer: B
A. Incorrect: Forming RNA primers is the role of RNA primase, not DNA polymerase I.
B. Correct: DNA polymerase I plays a key role in DNA replication by replacing the RNA primer at the start if each Okazaki fragment with a DNA strand.
C. Incorrect: The continuous synthesis of the leading strand is primarily carried out by DNA polymerase III, not DNA polymerase I.
D. Incorrect: Okazaki fragments are synthesized by DNA polymerase III on the lagging strand, while DNA polymerase I is involved in their processing after synthesis, particularly in removing primers.
Question 8
Which enzyme is associated with proofreading during DNA replication?
A. DNA primase
B. DNA helicase
C. DNA polymerase III
D. DNA ligase
Answer: C
A. Incorrect: DNA primase is responsible for synthesizing short RNA primers to initiate DNA replication, not for proofreading.
B. Incorrect: DNA helicase unwinds the DNA double helix to separate the two strands, making them available for replication, but it does not perform proofreading.
C. Correct: DNA polymerase III is the primary enzyme involved in DNA replication in bacteria and has a proofreading function. It can detect and correct errors by removing mismatched nucleotides using its `3'` to `5'` exonuclease activity.
D. Incorrect: DNA ligase joins the Okazaki fragments on the lagging strand by sealing nicks in the DNA backbone, but it does not have a proofreading role.
Question 9
(a) Explain why DNA must be replicated before mitosis and the role of helicase in DNA replication. [4]
(b) Explain how the base sequence of DNA is conserved during replication. [5]
(a) Any four of the following:
a. two genetically identical nuclei/daughter cells formed during mitosis (so hereditary information in DNA can be passed on);
b. two copies of each chromosome/DNA molecule/chromatid needed;
c. helicase unwinds the DNA/double helix;
d. to allow the strands to be separated;
e. helicase separates the two (complementary) strands of DNA;
f. by breaking hydrogen bonds between bases;
Sample answer:
DNA replication is essential before mitosis to ensure that each daughter cell receives an identical set of genetic information. During mitosis, a single cell divides into two genetically identical nuclei (daughter cells) [1]. For this to occur, each chromosome/DNA molecule/chromatid must be duplicated, providing two copies [1] - one for each daughter cell. Without replication, the daughter cells would lack the complete genetic material needed for proper function and heredity. Helicase is crucial as it unwinds the DNA double helix [1] by breaking the hydrogen bonds between the complementary base pairs [1]. This separation of the two strands allows them to serve as templates [1] for the synthesis of new DNA strands, initiating the replication process.
(b) Any five of the following:
a. DNA replication is semi-conservative;
b. DNA is split into two single/template strands;
c. nucleotides are assembled on/attached to each single/template strand;
d. by complementary base pairing;
e. adenine with thymine and cytosine with guanine / A with T and C with G;
f. strand newly formed on each template strand is identical to other template strand;
g. DNA polymerase used;
Sample answer:
The conservation of the DNA base sequence during replication is achieved through a semi-conservative mechanism [1], where each new DNA molecule consists of one original template strand and one newly synthesized strand. The process begins with the DNA molecule splitting into two single strands [1], onto which nucleotides are assembled or attached based on complementary base pairing rules: adenine pairs with thymine, and cytosine pairs with guanine [2]. This precise pairing ensures that the sequence of bases in the original strand is faithfully copied onto the new strand, resulting in a newly formed strand that is identical to the complementary original strand [1]. DNA polymerase is the key enzyme [1] that catalyzes this nucleotide addition.
Question 10
(a) Draw a labelled diagram of the structure of DNA, showing the arrangement of subunits. [3]
(b) Explain DNA replication. [5]
(a) Any three of the following:
a. correctly shows two antiparallel sugar-phosphate strands/backbones with linkages between phosphates and sugars connected through bases; (phosphate and simple names such as sugar and base are acceptable labels. They must be given at least once.)
b. correctly labeled phosphate and deoxyribose and base;
c. sugar linked to phosphates through correct pentagon corners/(`5’–3’`) linkages;
d. shows complementary base pairs of A-T/Adenine–Thymine and G-C/Guanine Cytosine;
e. correctly indicates both covalent/phosphodiester and hydrogen bonds;
Sample answer:
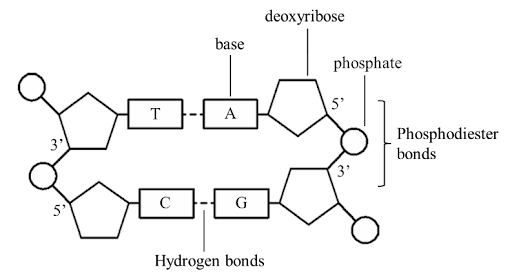
This diagram shows two antiparallel sugar-phosphate strands/backbones with linkages between phosphates and sugars connected through bases [1]. The diagram also correctly label phosphate, deoxyribose, and base, with the sugar (deoxyribose) linked to phosphates through the correct pentagon corners (`5'`to `3'`linkages) [2]. Additionally, the diagram shows complementary base pairs: adenine (A) paired with thymine (T) via two hydrogen bonds, and guanine (G) paired with cytosine (C) via hydrogen bonds; the backbones are linked by covalent phosphodiester bonds [2].
(b) Any three of the following:
a. DNA replication is semi-conservative/daughter DNA molecule contains one parent strand and one new strand;
b. unwinding of double helix/separation of two strands by helicase;
c. separated (parent) strands become templates for new strands;
d. free/single nucleotides join (parent/template) strands through complementary base pairing;
e. DNA polymerase joins nucleotides in new strands;
Sample answer:
DNA replication is a semi-conservative process, meaning that each daughter DNA molecule contains one parent strand and one newly synthesized strand [1], ensuring the accurate transmission of genetic information. The process begins with the unwinding of the double helix, where helicase plays a crucial role by separating the two strands through the breaking of hydrogen bonds between base pairs, creating a replication fork [1]. These separated parent strands then serve as templates for the synthesis of new strands, with the single-stranded DNA providing a template for replication [1]. Free nucleotides, present in the cellular environment, join the parent/template strands through complementary base pairing [1] - adenine pairing with thymine and guanine with cytosine -guided by the specific sequence of the original strand. DNA polymerase, a key enzyme in this process, facilitates the joining of these nucleotides into the new strands [1], adding them in a `5'` to `3'`direction and proofreading to maintain fidelity, resulting in two identical double-stranded DNA molecules, each retaining one original strand and one new strand.
Question 1
How can fragments of DNA be separated?
A. Using polymerase chain reaction (PCR)
B. Using gel electrophoresis
C. Using gene transfer
D. Using gene cloning
Question 2
Which technique causes fragments of DNA to move in an electric field?
A. Polymerase chain reaction (PCR)
B. Genetic modification
C. Therapeutic cloning
D. Gel electrophoresis
Question 3
What is amplified using the polymerase chain reaction (PCR)?
A. Large amounts of RNA
B. Small amounts of DNA
C. Small amounts of protein
D. Large amounts of polymers.
Question 4
What happens to DNA fragments in electrophoresis?
A. They move in a magnetic field and are separated according to their size.
B. They move in an electric field and are separated according to their size.
C. They move in a magnetic field and are separated according to their bases.
B. They move in an electric field and are separated according to their bases.
Question 5
How can forensic scientists obtain sufficient data from one hair follicle to make a reliable identification by DNA profiling?
A. By performing PCR with DNA from the sample
B. By digesting the sample with more than one restriction enzyme
C. By performing electrophoresis with many other known samples
D. By choosing a hair follicle with a particularly long hair
Question 6
The diagram shows a gel electrophoresis apparatus.

What can be deduced from the diagram?
A. X is the smallest fragment.
B. Y is positively charged.
C. X is moving towards the cathode.
D. Y travels at the greatest rate.
Question 7
What is the function of DNA polymerase I in DNA replication?
A. It forms primers by adding short lengths of RNA to the template strand.
B. It removes RNA primers and replaces them with DNA.
C. It builds the leading strand by adding DNA nucleotides continuously.
D. It forms Okazaki fragments by adding DNA nucleotides on the lagging strand.
Question 8
Which enzyme is associated with proofreading during DNA replication?
A. DNA primase
B. DNA helicase
C. DNA polymerase III
D. DNA ligase
Question 9
(a) Explain why DNA must be replicated before mitosis and the role of helicase in DNA replication. [4]
(b) Explain how the base sequence of DNA is conserved during replication. [5]
Question 10
(a) Draw a labelled diagram of the structure of DNA, showing the arrangement of subunits. [3]
(b) Explain DNA replication. [5]